Customer testimonials
Dr. Joshua Reyniers, Biochemistry, University of Sussex, Brighton, United Kingdom. "The Object Analyser has been incredibly helpful, the thresholding is considerably better than anything I can do in Fiji."
Dr. Cameron Nowell, Microscopy & Flow Cytometry Manager, Monash institute of Pharmaceutical Sciences, Melbourne, Australia. "Huygens has been running great for us and has become an integral part of a lot of our workflows, especially for iSIM and hyperspectral widefield imaging. We will definitely keep the Huygens Everywhere licence active."
Dr. Urs Ziegler, Director at the Center for Microscopy and Image Analysis at the University Zurich. "You (all at SVI) are doing a fantastic job, and the product is outstanding. During the (EMBL) course we had one of these datasets with a SNR of max 2 and the restoration of signals was fantastic."
Dr. Dr Neftali Flores Rodriguez, Sydney Microscopy & Microanalysis, University of Sydney, Australia."We found Huygens Object Analyzer to be the most robust and accurate solution for 3D colocalisation analysis (intracellular vesicles) compared to other software."
Dr. Nadia Halidi, Head of Advanced Light Microscopy Unit, CRG - Centre for Genomic Regulation, Barcelona, Spain."I truly appreciate your work and contributions to both the imaging community and QUAREP-LiMi. You and your team are doing a fantastic job."
Dr. Alexander Jurkevich - Director, Advanced Light Microscopy Core, University of Missouri, USA."I want to thank you and your colleagues at SVI for the fantastic support over the years. I am also proud that I somewhat contributed to the improvement of Huygens by being a beta tester of one of the earlier versions of the software."
Dr. Kunwar Jung KC, Biomedicine - Structural biology and drug discovery, University of Bergen, Norway. "I would like to express my sincere gratitude for your assistance with the deconvolution of the Dragonfly image, I truly appreciate the time and effort you dedicated to helping me, and I’m really pleased with the deconvolution on the Dragonfly image, it looks much sharper and better than the original."
Dr. Anastasiya Klebanovych, Advanced Bioimaging Laboratory, Donald Danforth Plant Science Center, Saint Louis, MO, USA. "We appreciate your support and the superb product you offer!"
Dr. Mathieu Catala, Microscopy Core, University of Sherbrooke, Québec, Canada. "I tried to stitch a file that has been shading corrected and it works flawlessly. Huygens Stitcher will be our option for all of our difficult tiles."
Matthias Klumpp, Institute for Physiology and Pathophysiology - Neurophysiology, University of Heidelberg, Germany. "We are very happy, and I especially love the new contrast options in Huygens 23.10 in the Twin Slicer and elsewhere, those are very nice quality of life improvements (especially with our dimly-lit fine structures).The Twin Slicer is such a simple tool. As soon as you have multichannel images or want to compare differently processed images, it becomes super useful."
Dr. Nicolai Urban, Light Microscopy Core, Max Planck - Florida Institute for Neuroscience, FL, USA. "Thank you very much for your help so far - I am very impressed with basically everyone from Huygens I have interacted with so far. Your user support is great!"
Dr. Austin Ferro, Neurobiology, Cheadle Laboratory, Cold Spring Harbor Laboratory, USA. "We have been so pleased with the Huygens so far and would love a quote to extend the license."
Dr. A. Phillip West, Microbial Pathogenesis and Immunology - College of Medicine, Texas A&M University, USA. "We are very happy with Huygens for our widefield images. My lab will is planning a lot more confocal so we are looking forward to using Huygens to deconvolve those pictures as well."
Prof. Ola Söderberg, Department of Pharmaceutical Biosciences, Uppsala University, Sweden. "Thanks for your immediate support! It is greatly appreciated!"
Dr. Aaron Barnes, Department of Microbiology, University of Minnesota, USA. "Huygens allows me to do my work - period. I work at a tough biological interface: bacterial interactions in microcolonies on the surface of host tissues (gut, the endovascular system, etc). And the bug I study most is *tiny* - you can fit more than a dozen of them inside an E. coli cell. Even using super-resolution techniques, it can be hard to demonstrate these interactions clearly. Using Huygens is like putting on my glasses in the morning - what was blurry and indistinct become clear and focused. Wins my personal "most valuable player" award every year. ;-)"
Prof. David Stone, Department of Biological Sciences, University of Illinois at Chicago, USA. "You have been a great help to us. I would recommend you and your company to anyone in need of your services."
Dr. Ioannis Alexopoulos, Microscopy & Image Analysis Specialist, General Instrumentation, Radboud University Nijmegen, The Netherlands. "I am stitching and deconvolving 600 2Ch-xy tiles (total size 160GB) and everything seems to work fine. Positioning of tiles happened using GPU, as well as deconvolving. Everything seems nice!"
Dr. Declan James, Department of Biochemistry, University of Wisconsin-Madison, USA "Thank you for all your help on the phone and for putting together a walk thru for PSF distiller (PALM/STORM) and use in Localizer"
Dr. Michael Paffett, Research Facility Director, Fluorescence Microscopy Shared Resource, University of New Mexico Cancer Center, USA "We used Huygens Localizer and were very impressed. The PC/GPU we built mowed through 30,000 frame in under 60s. Pretty bad-ass"
Dr. Shaun Walters, Research Facility Manager, School of Biomedical Sciences, The University of Queensland, Australia "Thank you so much! I really appreciate the support you and your company have provided us at UQ. "
Sven Hildebrand, Department of Cognitive Neuroscience, Maastricht Brain Imaging Centre (MBIC), Faculty of Psychology & Neuroscience, Maastricht University, The Netherlands "diSPIM Light Sheet images of DiI-labelled human brain tissue cleared with our newly published hFRUIT method, were considerably improved with Huygens de-skewing and deconvolution. Huygens 3D rendering allowed us to create awesome figures and videos, which really helped to let our data shine! "
Farhana Chowdhury, PhD student, Regenerative Medicine - Neural Tissue Engineering, Keele University, UK "I've tested the software and this seems to be very useful. I am a PhD student at the final stages and your software has helped clear some problems that I have been trying to fix for a while!"
Dr. Peter March, Bioimaging facility, Faculty of Biology, Medicine and Health, University of Manchester, UK "I have to say it’s great that SVI was so quick to help out at the beginning of shutdown. It’s definitely how I, and I know other people that use Huygens, felt about SVIs response at the start of the Covid-19 shutdown."
Dr. Petra Bolte, Core Facility Fluorescence Microscopy, University of Oldenburg, Germany, about Huygens@Home during COVID-19 crisis 'I really appreciate the service of SVI-Huygens to provide users with home licenses during the corona crisis. Thanks a lot!
Dr. Muriel Voisin, NUI Galway, Ireland, about Huygens@Home during COVID-19 crisis 'Thank you all guys for helping the rest of the community!"
Dr. Ute Kuhlicke, Heimholtz-Zentrum fuer Umweltforschung GmbH-UFZ, about Huygens@Home during COVID-19 crisis. 'Thanks a lot for this uncomplicated help in my work these days!"
Olivia Harding, Cellular & Molecular Biology, University of Pennsylvania, about Huygens@Home during COVID-19 crisis 'Thank you again for being so responsive during this difficult time!"
Merel Stiekema, Department of Molecular Cell Biology, University of Maastricht, The Netherlands 'I previously used ImageJ for Colocalization Analysis, but I actually find Huygens much more convenient and easier to use, moreover, it is much faster and the results are more consistent (i.e. a smaller standard deviation)"
Tom Pettini, Faculty of Biology, Medicine and Health, University of Manchester, UK, about Huygens@Home during COVID-19 crisis ''Many thanks for your quick reply with the licenses, and thanks for making these available, it has really helped me to be able to continue working on my images while University is closed!"
Prof. Rafael Garcia-Mata, (Harold C. and Charlotte L. Shaffer Chair in Biological Sciences) Department of Biological Sciences, University of Toledo, OH, USA. ''I’ve been teaching an Advance Microscopy Class and we got some spectacular results using Huygens"
Prof. Miguel Angel Peñalva Soto (CSIC Professor, EMBO member) Centro de Investigaciones Biológicas, Madrid, Spain. ''Everything is working smoothly. The colocalisation module is very good, the best I have tested so far"
Dr. Glyn Nelson, BioImaging Unit, campus for Ageing and Vitality, Newcastle University, UK "....we are quite impressed with the Huygens Array Detector results.''
Dr. Benjamin Lopez (NRI-MCDB Microscopy Facility Director, University of California Santa Barbara, USA "I tested the lightsheet fusion with 8-view lightsheet stacks of a drosophila ovary. The fusion worked great! What was especially useful was that it did the registration and fusion without fiducial marker beads in the images.''
Dr. Jürgen Schmied, CEO, GATTAquant (www.gattaquant.com), Germany "It’s always a pleasure to collaborate with SVI. There are not many collaborations getting done as smooth and straight forward as the ones with you."
Claudia Farb, Center for Neural Science at New York University, USA "Finally, your customer service and response time, from such knowledgeable people, are superb and unparalleled in today's world of cutbacks in customer service (I've been in the business for 30+ years). I really can't say enough wonderful things about Huygens and how happy we have been with its software"
Verónica Labrador Cantarero, MSc., Microscopy Unit, CNIC, Madrid, Spain ''Regarding customer service here at our Unit we only have good words for SVI and for you so thank you so much for your help!"
Dr. Lampros Panagis, Regeneron Pharmaceuticals Inc, USA "Thank you for the excellent support! I did not expect a phone call from Holland to discuss my application or my emails replied back in almost real time from the other side of the Atlantic; definitely not in August! We sometimes don’t get this kind of support from vendors that are based on the Pacific side of the continent, so this was definitely a huge plus for your software!"
Dr. Edward K. Williamson, Division of Cell Pathology, The Children’s Hospital of Philadelphia, USA "I just wanted to thank you for all the help you gave my division while we were deciding about purchasing Huygens. In fact, everyone at SVI were very helpful to me at every stage of the process." (June 2018)
Huong Ha, Neuroscience Program, Stanford University, USA "So far Huygens seems to surpass our other products at least for deconvolution and also surface rendering, and most of all, the technical support that you have provided is truly tremendous." (April 2018)
Dr. Christina Schlatterer, SFB969/LS Deuerling, University of Konstanz, Germany: "People in our lab who do confocal microscopy (and also those from other groups who use it) are really impressed by the program! We find it very important for our research with yeast cells and bacteria" (March 2017)
Dr. Priyam Banerjee, MD Anderson Cancer Center, Houston, Texas, USA: "I have been a Huygens user for 8-9 years now and the support provided by you guys are fantastic!'' (January 2017)"
Dr. Howard Berg, Integrated Microscopy Facility, Danforth Plant Science Center, St. Louis, USA: "SVI has proven to be the best for both customer service and online resources." (January 2017)''
Dr. Luke Hammond, Advanced Microscopy facility Queensland Brain Institute, Australia: "We are increasingly converting people over to deconvolution, especially for high-resolution laser scanning and spinning disk confocal projects. it’s been great to see how Huygens has made possible several projects that would otherwise have been very challenging or impossible. Having access to Huygens and the quick and experienced support from SVI gives staff at QBI a strong advantage in their microscopy based research." (September 2016)''
Dr. Jing Yan, Molecular Biology Labs, Princeton University, USA: "I want to express my deep appreciation of your great support!" (September 2015)
Dr. Shuoshuo Wang from the Weizmann Institute of Science, Israel: "I thank you all for providing such as wonderful tool that enriching our knowledge by revealing important detail information in cell biology.'' (May 2015)
Dr. Stefan Stanciu, Center for Microscopy-Microanalysis and Information Processing, University Politehnica of Bucharest, Romania: "I've been working with Huygens for couple of years now, and I can say that it's a really powerful tool. SVI have done a great job at developing a software package that is both flexible and user friendly, which is not always easy when it comes to scientific data processing. Additionally, the support I got was excellent. It is not only that SVI's team replies very fast to every inquiry, but they put consistent efforts into finding solutions for specific needs, which is really great." (February 2015)
Dr. Maral Tajerian, Department of Anesthesia, Stanford University, USA: "You guys have the best customer service I've ever seen!" (February 2015)
Dr. Jason Miller, Kellogg Eye Center, University of Michigan, USA: "This is the best experience I've ever had interacting with a scientific company - both in terms of patience and expertise." (February 2015)
Mahlon Collins, Center for Neuroscience, University of Pittsburgh, Pennsylvania, USA "We really appreciate your help and the flexibility in meeting our needs for the software." (October 2014)
Ikuroh OHSAWA, PhD., Biological Process of Aging, Tokyo Metropolitan Institute of Gerontology, Tokyo, Japan: "We enjoy your excellent software, and hope to extend the license. Thank you for your kind help!" (August 2014)
Dr. Stanislav Vinopal, AG Bradke (Axonal growth and regeneration), Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE), Bonn, Germany: "Huygens is an amazing piece of software and the way you approach and help your customers is great. It really deserves to be called real customer support!" (July 2014)
Dr. Mathias Pasche, LMB Light Microscopy Facility, MRC Laboratory of Molecular Biology (LMB), Cambridge, UK : "Overall, I think I haven't learned so efficiently for quite a while. Due to style of presentations, hands-on, small group and quality of presenters. Excellent! Thanks for the good time! Enjoyed your course from basics to scripting massively!"(May 2014)
Say-Tar Goh (Graduate student), Department of Biology, California Institute of Technology (Caltech), Pasadena USA: "So far Huygens worked fabulously for us - even in the earliest stage it was invaluable for calibrating and adjusting our microscope, so thanks!" (January 2014)
Dr. David Grunwald, Biochemistry and Molecular Pharmacology dept. - University of Massachusetts, USA: "With my colleagues I did "talk about our Huygens experience and the outstanding value we find in your support." (January 2014)
Dr. Dominic Waithe, Wolfson Imaging Facility, Weatherall Institute of Molecular Medicine - University of Oxford, UK: "The deconvolution and chromatic shift corrector have improved our bead representation. We are setting up an experiment where we are measuring the distance between points in 3D, visualised in different colour channels. The chromatic shift was a problem with our objective but now it seems much reduced, so we are happy." (November 2013)
Dr. Elisa De Luca, Center for Biomolecular Nanotechnologies @UNILE, Instituto Italiano di Tecnologia, Arnesano, Italy: "The course was much more interesting than I expected. I was positively surprised by the good preparation of the presenters and their ability of transmitting the key concepts - even to those who do not have any experience with deconvolution. Hands-on session was incredibly useful!" (October 2013)
Dr. Xiao-Dong Wang (Institute of Mental Health, Peking University, Beijing, China): "I am very happy with your software, and use it frequently now. Thanks for the excellent support from you and your colleagues!" (June 2013)
Prof. Benjamin Glick (Department of Molecular Genetics and Cell Biology, The University of Chicago, USA): "Thanks very much for the informative message! The results you provided are impressive." (February 2013)
MSc. Di Zi (Leiden Institute of Advanced Computer Science (LIACS), Leiden University, The Netherlands): "Our images are improved amazingly by the Huygens software. That's why we decided to buy the wide-field microscope deconvolution option, and we are very happy with it! Now we are thinking of buying the PSF distill option as well. "(February 2013)
Dr. Zoltan Cseresnyes, (Confocal and 2-Photon Microscopy Core Facility - MCF - Max Delbrueck Center for Molecular Medicine, Berlin, Germany): "Both my facility users and I are very impressed with the new (and not so new) features, and some of my new users are really enjoying the object analyzer and the ROI options!" (August 2011)
Dr. Pete Bankhead, (Bio Quant - Nikon Imaging Centre - Heidelberg, Germany): " s for comments, I would like to be clear that the more I use the HRM, the more I like it! I think it's excellent for general use, and the interface is very easy for new users to learn. So you should know that I am impressed by it, and I really appreciate your help in getting it working well for us." (July 2011)
Dr. Shantanu Sur, (Institute for Bio Nano Technology in Medicine, Northwestern University, Chicago, USA): "We are also happy to renew the software agreement. Thanks for the warm support as always. We have got some nice images rendered by Huygens." (June 2011)
Dr. James Burchfield, (Molecular Imaging Facility, Garvan Institute of Medical Research, The University of New South Wales, Australia): "The speed and fluidity of the program is outstanding. I am also very impressed by the output from the alignment tool." (June 2011)
MSc. Johannes Koch, (Department of Biochemistry and Cell Biology, MFPL, University of Vienna, Austria): "I am convinced that not only the scientifically sound deconvolution and analysis software but also the excellent and personal support provided by Huygens will make Huygens the software of choice for scientific image processing." (October 2010)
"Dr. Thomas Launey (BSI: www.brain.riken.jp):" "Dr. Launey is a Unit Leader at the RIKEN Brain Science Instituein Wako-shi, Japan. His team explores the cellular and molecular mechanisms of motor learning in the cerebellum. Studying neural networks involves visualizing the dynamic behaviors of molecules and cells in the live brain. Dr. Launey: “We use Huygens to study receptor distribution and movement: the receptors are tagged with fluorescent protein, Qdots or organic dyes and imaged at high resolution. Without deconvolution we can't really resolve the position of the receptors since they moves within a dendrite which is full of small protrusions called spines. Without deconvolution we can't really assess position within the spine or co-localization with other proteins for example.” In the future, Dr. Launey plan to make full use of the new particle tracking function of Huygens and use correlative optical/electron microscopy to further enhance resolution."(June 2010)
Dr. Manuel Izquierdo Pastor, (CSIC-Universidad Autonoma de Madrid- Facultad de Medicina, Spain): "Huygens features and the superb support you guys provide are important reasons why we bought Huygens. In addition, your on-line support encourages all-level users to face deconvolution and also helps a lot to avoid computer-artifacts, to double check achieved results, etc.." (September 2009)
MSc. Daniel Veilleux (École Polytechnique de Montréal, Québec Canada): "Indeed, the numerous information provided by SVI was extremely useful in solving our issues with confocal imaging and deconvolution. We were able to identify a problem with the z-stepper of our microscope and bring about the required temporary modifications to correct for its deficiency while expecting a routine maintenance exercise to come. With these corrections and the default PSF, our images now have a relative error of less than 3-5% in the z-axis, compared to a previous deformation of 300-400%..." "Thank you very much for the great help you've provided." (March 2009)
Dr. Jonathan Foley (Bio-engineering, University of California Berkeley, USA): "We are running Huygens Core in a completely headless environment on our cluster. We have developed custom scripts that generate deconvolution jobs and then hand them off to the Sun Grid Engine scheduler. Its working out great. Our analysis pipeline is an order of magnitude faster now. We have Huygens Core running on 8 (Dual-Opteron 2Gb RAM) nodes. It takes 4 days to deconvolve 84 fields of 3 channel x 86 slices x1024x1024. Our datasets are pretty big because we are collecting statistics on a large number of cells. Previously, running Huygens Pro on a Mac Pro where we could only use one or two processors this took a month!!!" (January 2009)
Dr. Juraj Kabat (Biological Imaging Facility, NIH/NIAID, Bethesda, USA): "We enjoy the new features/improvements in Huygens 3.3. This is the best release so far. Also twin slicer improvements are significant." (December 2008)
Dr. Michael Paddy (Section of Molecular and Cellular Biology, University of California - Davis, USA): "I thought you would be interested in this very dramatic difference when using the Huygens vs another decon package for noisy, live data. In theory, the Huygens should be better for noisy data like this because of the method it uses to generate the image updates in the decon iteration, but I was surprised to see just how much better it is." (2007)
Dr. James Evans (MIT CSBi, USA): "Because software companies generally respond to researchers' needs, most current imaging limitations occur downstream of acquisition", explains James G. Evans, research scientist at the Computational and Systems Biology Initiative (CSBi).
Evans and colleagues are developing an imaging pipeline to collect data directly from microscopes and shuttle this data at high speeds into a database for storage and management, where the data can then be sent for image restoration by deconvolution. To this end, Evans says he has had a lot of success with SVI's Huygens software. "It's very well parallelized, working across many processors, and is almost platform-independent," Evans says. (ref.: MIT CSBi).
Dr. Sergei Bushuk (Stepanov Institute of Physics, Minsk, Belarus): "I'm 100% happy with the software. In fact, Huygens proved to be highly effective (and cost effective) solution. With correctly measured stacks the results are impressing."
Louis Villeneuve (Research Associate - Confocal Microscopy Heart Montreal Institute - Research Center, Canada): "Thanks for the new version. I really like your product, Huygens Pro is very complete and accurate."
Dr. Ray Gilbert (University of Auckland, Bio - Engineering Department, New Zealand): "This is the software that we ended up purchasing, after trialling with two other packages. I found it to be the easiest to use of the 3 packages and their support was excellent (we had a few hiccups initially and they kept extending the free license for about 3 months while I tested everything I needed)."(Jan 2008)
Dr. Jakub Sikora (Charles University, 1rst Faculty of Medicine, Prague, Czech Republic): "SVI is one of the least commercial commercial companies that I know." (March 2007)
Dr. Rolly Wiegand (Head CALM facility University of Edingburgh, UK): "To make it short, we have very good experiences with the Huygens deconvolution software from SVI (no commercial interest here). The recent upgrade of the software gives you some convenient batching utilities. However, if you try Huygens Script in addition you will be able to write your own macros for batching and a lot of other applications. This might be a bit tricky at the beginning, but is worth the time spent. My recommendation is to run the software on sufficiently powerful workstations, preferably dual CPU machines with a minimum of 4GB of RAM. Just talk to the SVI guys and have a look at their website/knowledge base and try a test version on your own hardware." (Confocal list server, July 2006)
Dr. Andrew Resnick (Case Western Reserve University - Physiology and Biophysics, USA): "The Huygens Deconvolution Software just makes the images look better. After deconvolution the images are cleaned up (the noise is removed, more details in the image). It is much easier to make a statement with deconvolved images."
Dr. Ulrike Engel (Scientific Director of the NIC Imaging Centre, Bioquant, University of Heidelberg, Germany): "Researchers who use Huygens at the facility to enhance their confocal data are very much impressed by the results. I would not have anticipated myself how much especially noisy confocal data can be improved by deconvolution with Huygens. As noisy data is the rule rather than the exception with live samples, we now use Huygens routinely to enhance our spinning disk data."
"At our Annual Fluorescence Microscopy Course Live Imaging in 3D course the short training in deconvolution with Huygens was very popular with students and had an addictive quality. Students came back the next day to deconvolve more image stacks. "
S. Kumar, G. Ho, K.M. Woo and L. Zhuo (Institute of Bioengineering and Nanotechnology, Singapore): "Estimating the underlying fluorescence signal: Figure 1(a) is a typical image of the optic disc in the retina of a transgenic mouse where the fluorescence signal is from the GFP protein which, in this case, labels the glial cells. As observed, the image has a very low SNR and this makes it extremely difficult to detect any suitable landmark points and it is clearly inappropriate to use such images as reference images in rank matching. An a-priori estimate is therefore required to accurately estimate the underlying fluorescence signal. We have explored several methods of determining this estimate and found that the Huygens deconvolution method (Huygens Essential Software, Scientific Volume Imaging BV) gives the best results. The deconvolved result of Fig. 1(a) is shown in Fig. 1(b) and as observed, the speckle noise is removed and the fluorescence intensity profile is characterized by various intensity maxima. Conversely, a 9 × 9 median filtering method of estimating the a-priori estimate fails to remove the speckle noise and instead distorts the image as observed in Fig. 1(c). Figure 1(d) shows the results of applying anisotropic diffusion where the noise appears to be removed but at the expense of diffusing the underlying fluorescence signal resulting in the image appearing blurred overall."
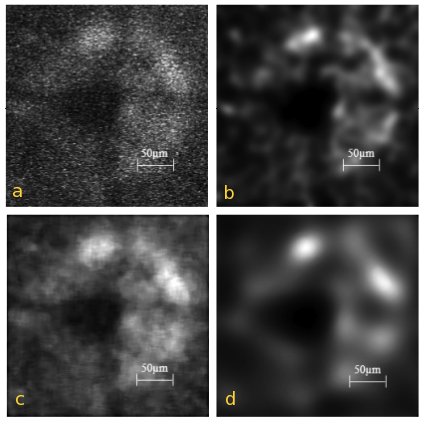
Fig. 1. (a) Input image, of the GFP signal from the optic disc of the transgenic mouse. (b)–(d) restored image obtained after subjecting the input image to (b) Huygens deconvolution (c) 9 × 9 median filtering and (d) speckle reducing anisotropic diffusion.
References to SVI software
Quite some scientific papers mention SVI's Software. See a brief list in Huygens References.

Attendants to courses and training-sessions
SVI gives training sessions at our office at The Netherlands on a regular basis. We also give workshops and courses in conferences and institutions all over the world. (For more information see TrainingAndCourses). Next to that we also offer for our customers and their users remote tailor made training sessions as part of the M&U services. These are a few attendants' opinions about our courses:
After a two day course at the NIH, Bethesda, USA:
- ...we were very impressed by the presentations. He really managed to get us into what deconvolution can do for our images and to explain all the complicated
algorithms without showing one mathematical equation. This was absolutely brilliant. ... This workshop was excellent.
- ...able to explain physics concepts without using algorithms. This is rare. He gave a small philosophical introduction on reality and truth, which set the tone perfectly for his explanation of image restoration and deconvolution. It was very motivating to hear the explanations on the software.
- ...a very informative course!!!
After a Huygens Professional workshop at our office:
- ...the course has been great!!! I'm also much more confident with the Huygens Pro. The tcl command window is really powerful.
- Thank you again for the hospitality and the well organized course!!!
- The workshop gave insight into the manifold of possibilities provided by Huygens. I really enjoyed the final session, where we could discuss our individual problems and scientific question with the developers.
After a Huygens deconvolution workshop at our office:
- I enjoyed the course a lot and profited from it!!! The level of the audience was very high and we had a lot of stimulating discussions.
- Dr. Pawel Pasierbek (IMP-IMBA, Vienna):
This was a highly recommendable course with the aim to help us to optimize the usage of the SVI decon software. I would suggest to try the software beforehand in order to be able to use expertise of Hans, Jose and Edwin during the course. Bring your difficult data along with the results of your decon attempts, and discuss what can be done better.
