Blur removal and resolution boost in conventional microscopic images
Total blur removal in conventional microscopic images is realized in the Huygens Software by reconstructing information which was lost in the image formation process.
In the left picture in the image pair below a cooled CCD recorded image of a bladder cell nucleus is shown as rendered by the Simulated Fluorescence Process algorithm:


The picture on the right shows the result after applying the Maximum Likelihood Estimation restoration algorithm incorporated in the Huygens System. This introduces a virtual optical sectioning effect not unlike the (physical) sectioning effect in the Confocal Microscope. This dramatically reduces the non-specific labeled fluorescence signal of the nucleus, whereas the labeled centromeres become clearly visible. Image recorded by Hans Vrolijk, Willem C.R. Sloos and Jan Slats.
The resolution boost is clearly visible in the X-Z section shown below. The original data is displayed in red: the cone-like spreading of the out of focus light from the centromeres can be clearly seen. The restored image is displayed in white. Since the section does not pass right through the center of the left centromere, the resolution improvement introduced by the Silicon Slicer causes it to be invisible in the restored (white) image.
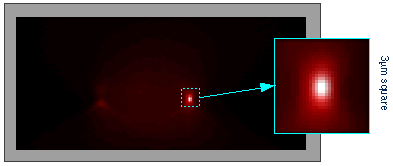
The inset shows the restored right centromere at high magnification. The axial size of the restored centromere image is below 0.5 µm: better than the axial resolution of a Confocal Microscope!!! But then, restoring confocal images may well turn the table...
A detailed case
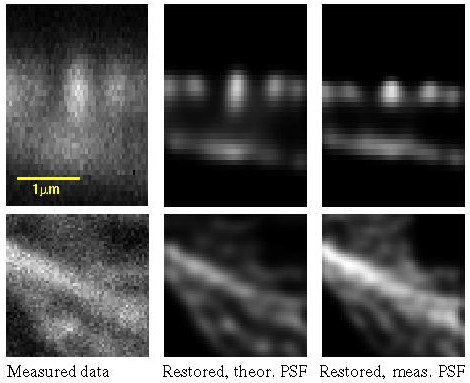
Actin filaments of mouse fibroblast cells as recorded with a two-photon Confocal Microscope and restored with MLE with theoretical and measured PSF. Maximum photon count: 204. Top row: XZ sections (y = 32); bottom row: XY sections (z = 11).
What follows is a SFP rendering of the restored image (bottom view).

Reliability of the result
After Image Restoration structures become visible which were not apparent in the original data: are they true object structures? To gain insight in these problems we conduct a series of experiments with synthetic data.
To analyze two-object resolution (see Rayleigh Criterion) a pseudo synthetic image was constructed in the following manner:
- The previous restored fibroblast image was soft-thresholded and clipped to generate a test object with small constant-intensity areas. A constant Background (5% of max) was added.
- Several small objects in the image were duplicated in both X- and Z- direction, at distances below unrestored resolution.
- The object was imaged using the measured Point Spread Function of the microscope (as explained in the Cookie Cutter).
- Photon Noise was added (200 photons in the brightest areas).
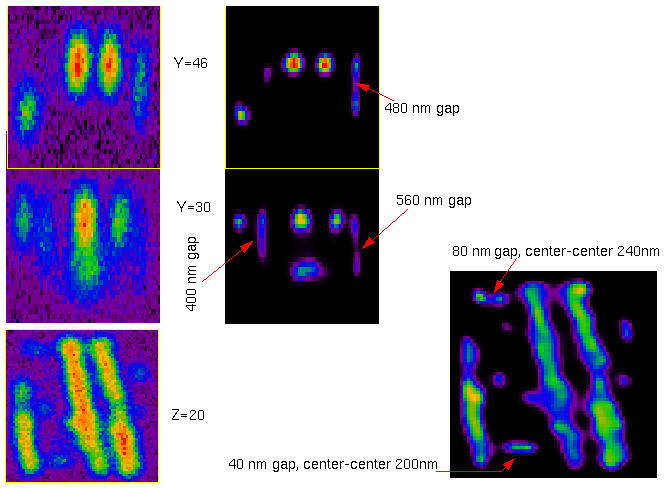
Synthetic test object derived from the restored mouse fibroblast image. Small objects are duplicated in both x- and z-direction to estimate two-object resolution:
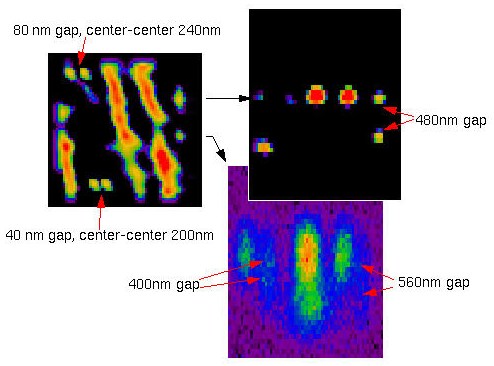
After imaging, points that are too close to each other appear as single objects.
After restoration, we can recover the two points separated 80 nm along X, but the 40 nm gap has disappeared. In the Z direction the resolution is also improved, until certain limits: a gap of 480 nm can still be recovered, but another of 400 nm remains totally bridged.
The limits
The resolution limits attainable with restored confocal microscopy have been explored in an experiment by Stefan W. Hell 1 in which reflecting gold particles were imaged with a high precision stage-scanning confocal microscope. The results show an axial resolution (FWHM) in the order of 100 nm and an lateral resolution in the order of 50 nm. These results set an upper limit as to what can be achieved with deconvolution of fluorescent images: in practice, fluorescence images have higher levels of Photon Noise that lower the resolution.
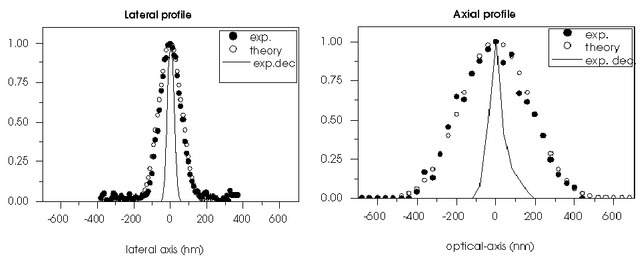
See also the FAQ "How large a resolution improvement can be expected from the Huygens deconvolution?".
More examples
Here below you can see another example. (Confocal data: isolated Rat Hepatocyte couplet recorded by Dr. Permsin Marbet at the Department of Anatomy, University of Basel, Switzerland, in the lab of Prof. Lukas Landmann).
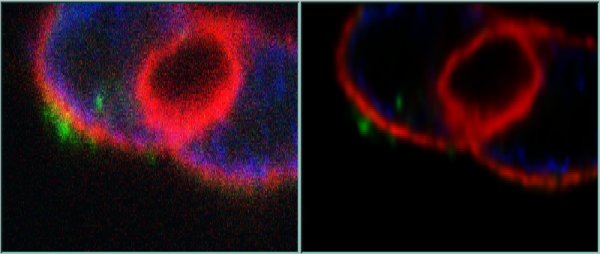 XZ slices, original and deconvolved with Huygens
XZ slices, original and deconvolved with Huygens
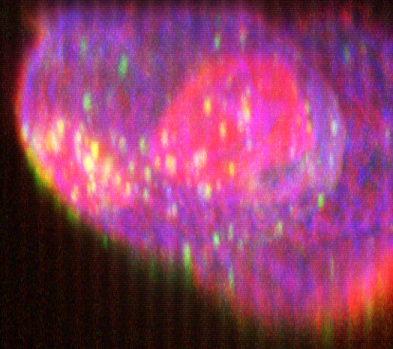
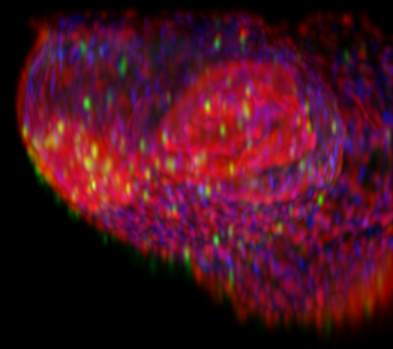 XZ Maximum Intensity Projection (MIP) made with the Fast Mip renderer, original and deconvolved with Huygens
XZ Maximum Intensity Projection (MIP) made with the Fast Mip renderer, original and deconvolved with Huygens
References
1 Schrader, M., S. W. Hell and H.T.M. van der Voort. (1996) Potential of confocal microscopes to resolve in the 50-100 nm range. Appl. Phys. Lett. 69 (24), pp. 3644-3646.
Search the FAQ for "resolution gain".
